3D Protein Structures Reveal Hidden Links in the Evolutionary Tree
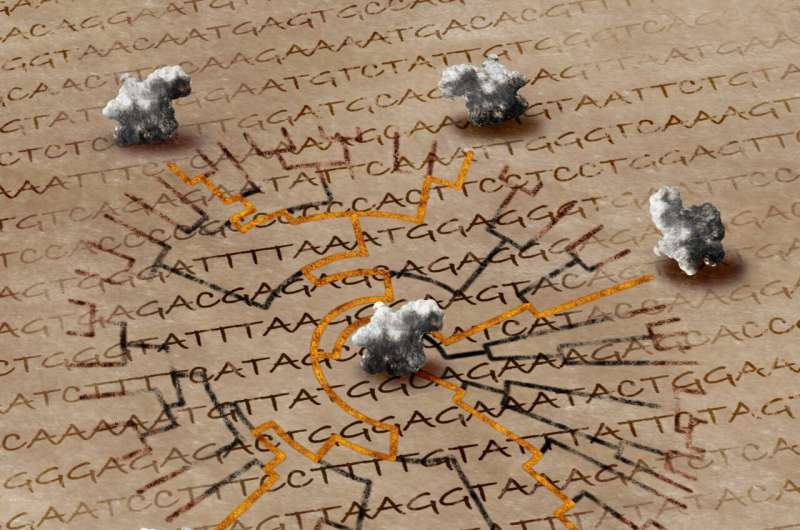
According to a study published in Nature Communications, the three-dimensional form of a protein can be utilized to determine long-standing evolutionary links in the tree of life.
The reliability of evolutionary trees, a vital tool used by the scientific community to comprehend the origins of life, track the spread of pathogens, or develop novel treatments for illness, has been enhanced for the first time by researchers combining data from protein shapes with data from genomic sequences.
Importantly, the method is effective even when proteins with predicted structures that have never been determined experimentally are used. It affects the enormous volume of structural data produced by programs such as AlphaFold 2 and provides new insights into the prehistoric past of life on earth.
The reliability of evolutionary trees, a vital tool used by the scientific community to comprehend the origins of life, track the spread of pathogens, or develop novel treatments for illness, has been enhanced for the first time by researchers combining data from protein shapes with data from genomic sequences.
Importantly, the method is effective even when proteins with predicted structures that have never been determined experimentally are used. It affects the enormous volume of structural data produced by programs such as AlphaFold 2 and provides new insights into the prehistoric past of life on earth.
Did you know? You can comment on this post! Just scroll down
While 250 million protein sequences are known, 210,000 protein structures have been determined experimentally. In the coming years, projects such as the EarthBioGenome project may produce billions more protein sequences. The volume of data makes it possible to use the method on a never-before-seen scale.
By tracking the ways in which species and genes diverge from common predecessors, researchers have been reconstructing evolution for many decades. In order to infer links, these evolutionary or phylogenetic trees are typically constructed by comparing DNA or protein sequences and calculating the similarities and differences.
However, a major obstacle that researchers must overcome is saturation. Signals suggesting a common ancestry can be erased when genetic sequences undergo such drastic changes over extended periods of time that they no longer resemble their ancestral forms.
By tracking the ways in which species and genes diverge from common predecessors, researchers have been reconstructing evolution for many decades. In order to infer links, these evolutionary or phylogenetic trees are typically constructed by comparing DNA or protein sequences and calculating the similarities and differences.
However, a major obstacle that researchers must overcome is saturation. Signals suggesting a common ancestry can be erased when genetic sequences undergo such drastic changes over extended periods of time that they no longer resemble their ancestral forms.
The study's principal author, Dr. Cedric Notredame, a researcher at the Center for Genomic Regulation (CRG), states that "the issue of saturation dominates phylogeny and represents the main obstacle for the reconstruction of ancient relationships." It resembles the deterioration of an old manuscript. The message is lost when the letters get hazy.
The study team looked to the physical architecture of proteins in order to overcome this obstacle. The intricate configurations that proteins fold into dictate the function of a cell. Compared to the sequences themselves, these forms are more preserved across evolutionary time, which means they change more slowly and hold onto ancestral traits for longer.
The amino acid sequence of a protein determines its structure. Even though sequences might change, the general structure usually stays the same to maintain functionality. The researchers thought that by measuring intra-molecular distances (IMDs), or the distance between pairs of amino acids within a protein, they could determine how much the structures differ with time.
A vast collection of proteins with known structures from a variety of animals was gathered for the study. In order to create phylogenetic trees, they determined the IMDs for every protein.
They discovered that while trees constructed from structural data closely resembled those obtained from genetic sequences, the structural trees had one significant advantage: they were less susceptible to saturation. This indicates that even after genetic sequences had diverged considerably, they were still able to detect consistent signals.
The study team looked to the physical architecture of proteins in order to overcome this obstacle. The intricate configurations that proteins fold into dictate the function of a cell. Compared to the sequences themselves, these forms are more preserved across evolutionary time, which means they change more slowly and hold onto ancestral traits for longer.
The amino acid sequence of a protein determines its structure. Even though sequences might change, the general structure usually stays the same to maintain functionality. The researchers thought that by measuring intra-molecular distances (IMDs), or the distance between pairs of amino acids within a protein, they could determine how much the structures differ with time.
A vast collection of proteins with known structures from a variety of animals was gathered for the study. In order to create phylogenetic trees, they determined the IMDs for every protein.
They discovered that while trees constructed from structural data closely resembled those obtained from genetic sequences, the structural trees had one significant advantage: they were less susceptible to saturation. This indicates that even after genetic sequences had diverged considerably, they were still able to detect consistent signals.
Understanding that both sequences and structures provide insightful information, the team created a hybrid strategy that enhanced the tree branches' dependability while also assisting in differentiating between associations that are right and those that are not.
"It's similar to having two witnesses recount an incident from different perspectives," says research co-author Dr. Leila Mansouri. "Each provides unique details, but together they give a fuller, more accurate account."
Understanding the connections between kinases in the human genome is one real-world instance where the combined approach could have a big influence. Proteins called kinases are engaged in a wide range of crucial cellular processes.
"It's similar to having two witnesses recount an incident from different perspectives," says research co-author Dr. Leila Mansouri. "Each provides unique details, but together they give a fuller, more accurate account."
Understanding the connections between kinases in the human genome is one real-world instance where the combined approach could have a big influence. Proteins called kinases are engaged in a wide range of crucial cellular processes.
"The genome of most mammals, including humans, contains about 500 protein kinases that regulate most aspects of our biology," according to Dr. Notredame. "These kinases are major targets for cancer therapy, for example drugs like imatinib for humans or toceranib for dogs."
Over the past billion years, duplications have given rise to human kinases. "Within the human genome, the most distantly related kinases are about a billion years apart," adds Professor Notredame. "They duplicated in the common ancestor of the common ancestor of our common ancestor."
Building precise gene trees that demonstrate the relationships between all of these kinases is extremely challenging due to the large timescale required.
Over the past billion years, duplications have given rise to human kinases. "Within the human genome, the most distantly related kinases are about a billion years apart," adds Professor Notredame. "They duplicated in the common ancestor of the common ancestor of our common ancestor."
Building precise gene trees that demonstrate the relationships between all of these kinases is extremely challenging due to the large timescale required.
"Despite its flaws, the kinase evolutionary tree is frequently utilized to comprehend how it interacts with other medications. Enhancing this tree or trees belonging to other significant protein families would be a significant step forward for human health, Dr. Notredame continues.
The work's potential uses extend beyond cancer. Using the method to produce more precise evolutionary trees may also help us better understand how diseases evolve in general, which could help with the creation of medicines and vaccinations.
They can also direct the development of novel enzymes for biotechnology, help uncover the origins of complex features, and even track the evolution of species in reaction to climate change.
The work's potential uses extend beyond cancer. Using the method to produce more precise evolutionary trees may also help us better understand how diseases evolve in general, which could help with the creation of medicines and vaccinations.
They can also direct the development of novel enzymes for biotechnology, help uncover the origins of complex features, and even track the evolution of species in reaction to climate change.
Article Posted 4 Months ago. You can post your own articles and it will be published for free.
No Registration is required! But we review before publishing! Click here to get started
One Favour Please! Subscribe To Our YouTube Channel!
468k
Cook Amazing Nigerian Dishes, Follow Adorable Kitchen YouTube Channel!
1.1m
Like us on Facebook, Follow on Twitter
React and Comment
Click Here To Hide More Posts Like This
Watch and Download Free Mobile Movies, Read entertainment news and reports, Download music and Upload your own For FREE.
Submit Your Content to be published for you FREE! We thrive on user-submitted content!
But we moderate!